Abstract
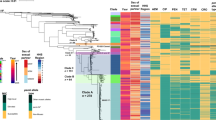
Chlamydia trachomatis is responsible for both trachoma and sexually transmitted infections, causing substantial morbidity and economic cost globally. Despite this, our knowledge of its population and evolutionary genetics is limited. Here we present a detailed phylogeny based on whole-genome sequencing of representative strains of C. trachomatis from both trachoma and lymphogranuloma venereum (LGV) biovars from temporally and geographically diverse sources. Our analysis shows that predicting phylogenetic structure using ompA, which is traditionally used to classify Chlamydia, is misleading because extensive recombination in this region masks any true relationships present. We show that in many instances, ompA is a chimera that can be exchanged in part or as a whole both within and between biovars. We also provide evidence for exchange of, and recombination within, the cryptic plasmid, which is another key diagnostic target. We used our phylogenetic framework to show how genetic exchange has manifested itself in ocular, urogenital and LGV C. trachomatis strains, including the epidemic LGV serotype L2b.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
World Health Organization, Department of Reproductive Health and Research. Prevalence and Incidence of Selected Sexually Transmitted Infections. (World Health Organization, Geneva, Swizerland, 2011).
Mariotti, S.P., Pascolini, D. & Rose-Nussbaumer, J. Trachoma: global magnitude of a preventable cause of blindness. Br. J. Ophthalmol. 93, 563–568 (2009).
Burgoyne, R.A. Lymphogranuloma venereum. Prim. Care 17, 153–157 (1990).
Behets, F.M. et al. Chancroid, primary syphilis, genital herpes, and lymphogranuloma venereum in Antananarivo, Madagascar. J. Infect. Dis. 180, 1382–1385 (1999).
Mabey, D. & Peeling, R.W. Lymphogranuloma venereum. Sex. Transm. Infect. 78, 90–92 (2002).
Viravan, C. et al. A prospective clinical and bacteriologic study of inguinal buboes in Thai men. Clin. Infect. Dis. 22, 233–239 (1996).
Clarke, I.N. Evolution of Chlamydia trachomatis. Ann. NY Acad. Sci. 1230, E11–E18 (2011).
Gaydos, C.A. Nucleic acid amplification tests for gonorrhea and Chlamydia: practice and applications. Infect. Dis. Clin. North Am. 19, 367–386, ix (2005).
Fredlund, H., Falk, L., Jurstrand, M. & Unemo, M. Molecular genetic methods for diagnosis and characterisation of Chlamydia trachomatis and Neisseria gonorrhoeae: impact on epidemiological surveillance and interventions. APMIS 112, 771–784 (2004).
Nunes, A., Borrego, M.J., Nunes, B., Florindo, C. & Gomes, J.P. Evolutionary dynamics of ompA, the gene encoding the Chlamydia trachomatis key antigen. J. Bacteriol. 191, 7182–7192 (2009).
Pudjiatmoko, Fukushi, H., Ochiai, Y., Yamaguchi, T. & Hirai, K. Phylogenetic analysis of the genus Chlamydia based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 47, 425–431 (1997).
Klint, M. et al. High-resolution genotyping of Chlamydia trachomatis strains by multilocus sequence analysis. J. Clin. Microbiol. 45, 1410–1414 (2007).
Pannekoek, Y. et al. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol. 8, 42 (2008).
Brunelle, B.W. & Sensabaugh, G.F. The ompA gene in Chlamydia trachomatis differs in phylogeny and rate of evolution from other regions of the genome. Infect. Immun. 74, 578–585 (2006).
Dean, D. et al. Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerg. Infect. Dis. 15, 1385–1394 (2009).
Seth-Smith, H.M. et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10, 239 (2009).
Jeffrey, B.M. et al. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect. Immun. 78, 2544–2553 (2010).
Joseph, S.J., Didelot, X., Gandhi, K., Dean, D. & Read, T.D. Interplay of recombination and selection in the genomes of Chlamydia trachomatis. Biol. Direct 6, 28 (2011).
Ikryannikova, L.N., Shkarupeta, M.M., Shitikov, E.A., Il'ina, E.N. & Govorun, V.M. Comparative evaluation of new typing schemes for urogenital Chlamydia trachomatis isolates. FEMS Immunol. Med. Microbiol. 59, 188–196 (2010).
Millman, K.L., Tavare, S. & Dean, D. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 183, 5997–6008 (2001).
Stephens, R.S. et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759 (1998).
Croucher, N.J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 (2011).
Stephens, R.S. Chlamydiae in evolution: a billion years and counting. in Proceedings of the Tenth International Symposium on Human Chlamydial Infections. (eds. Schachter, J. et al.) 3–12 (Antalya, Turkey, 2002).
Suchland, R.J., Eckert, L.O., Hawes, S.E. & Stamm, W.E. Longitudinal assessment of infecting serovars of Chlamydia trachomatis in Seattle public health clinics: 1988–1996. Sex. Transm. Dis. 30, 357–361 (2003).
Nieuwenhuis, R.F., Ossewaarde, J.M., van der Meijden, W.I. & Neumann, H.A. Unusual presentation of early lymphogranuloma venereum in an HIV-1 infected patient: effective treatment with 1 g azithromycin. Sex. Transm. Infect. 79, 453–455 (2003).
Hayes, L.J. et al. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect. Immun. 62, 5659–5663 (1994).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997).
LeQuesne, W. A method of selection of characters in numerical taxonomy. Syst. Biol. 18, 201–205 (1969).
Bruen, T.C., Philippe, H. & Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681 (2006).
Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 34, 126–129 (1992).
Jakobsen, I.B. & Easteal, S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput. Appl. Biosci. 12, 291–295 (1996).
Gomes, J.P. et al. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 17, 50–60 (2007).
Kuo, C.C., Wang, S.P., Grayston, J.T. & Alexander, E.R. TRIC type K, a new immunologic type of Chlamydia trachomatis. J. Immunol. 113, 591–596 (1974).
Unemo, M. & Clarke, I.N. The Swedish new variant of Chlamydia trachomatis. Curr. Opin. Infect. Dis. 24, 62–69 (2011).
Unemo, M. et al. The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization. Microbiology 156, 1394–1404 (2010).
Misyurina, O.Y. et al. Mutations in a 23S rRNA gene of Chlamydia trachomatis associated with resistance to macrolides. Antimicrob. Agents Chemother. 48, 1347–1349 (2004).
Jones, R.B., Van der Pol, B., Martin, D.H. & Shepard, M.K. Partial characterization of Chlamydia trachomatis isolates resistant to multiple antibiotics. J. Infect. Dis. 162, 1309–1315 (1990).
Lefevre, J.C., Lepargneur, J.P., Guion, D. & Bei, S. Tetracycline-resistant Chlamydia trachomatis in Toulouse, France. Pathol. Biol. (Paris) 45, 376–378 (1997).
Somani, J., Bhullar, V.B., Workowski, K.A., Farshy, C.E. & Black, C.M. Multiple drug-resistant Chlamydia trachomatis associated with clinical treatment failure. J. Infect. Dis. 181, 1421–1427 (2000).
Binet, R. & Maurelli, A.T. Fitness cost due to mutations in the 16S rRNA associated with spectinomycin resistance in Chlamydia psittaci 6BC. Antimicrob. Agents Chemother. 49, 4455–4464 (2005).
Brunham, R. et al. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Invest. 94, 458–463 (1994).
Gomes, J.P., Bruno, W.J., Borrego, M.J. & Dean, D. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 186, 4295–4306 (2004).
Jurstrand, M. et al. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 39, 3915–3919 (2001).
van Duynhoven, Y.T., Ossewaarde, J.M., Derksen-Nawrocki, R.P., van der Meijden, W.I. & van de Laar, M.J. Chlamydia trachomatis genotypes: correlation with clinical manifestations of infection and patients' characteristics. Clin. Infect. Dis. 26, 314–322 (1998).
Moncan, T., Eb, F. & Orfila, J. Monoclonal antibodies in serovar determination of 53 Chlamydia trachomatis isolates from Amiens, France. Res. Microbiol. 141, 695–701 (1990).
Wagenvoort, J.H., Suchland, R.J. & Stamm, W.E. Serovar distribution of urogenital Chlamydia trachomatis strains in The Netherlands. Genitourin. Med. 64, 159–161 (1988).
Barnes, R.C., Suchland, R.J., Wang, S.P., Kuo, C.C. & Stamm, W.E. Detection of multiple serovars of Chlamydia trachomatis in genital infections. J. Infect. Dis. 152, 985–989 (1985).
Hanna, L., Thygeson, P. & Jawetz, E. Elementary-body virus isolated from clinical trachoma in California. Science 130, 1339–1340 (1959).
Spaargaren, J. et al. Analysis of Chlamydia trachomatis serovar distribution changes in the Netherlands (1986–2002). Sex. Transm. Infect. 80, 151–152 (2004).
Dean, D. & Stephens, R.S. Identification of individual genotypes of Chlamydia trachomatis from experimentally mixed serovars and mixed infections among trachoma patients. J. Clin. Microbiol. 32, 1506–1510 (1994).
Machado, A.C. et al. Distribution of Chlamydia trachomatis genovars among youths and adults in Brazil. J. Med. Microbiol. 60, 472–476 (2011).
Batteiger, B.E. et al. Correlation of infecting serovar and local inflammation in genital chlamydial infections. J. Infect. Dis. 160, 332–336 (1989).
Millman, K. et al. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 186, 2457–2465 (2004).
Persson, K. & Osser, S. Lack of evidence of a relationship between genital symptoms, cervicitis and salpingitis and different serovars of Chlamydia trachomatis. Eur. J. Clin. Microbiol. Infect. Dis. 12, 195–199 (1993).
Lysén, M. et al. Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish County. J. Clin. Microbiol. 42, 1641–1647 (2004).
Sturm-Ramirez, K. et al. Molecular epidemiology of genital Chlamydia trachomatis infection in high-risk women in Senegal, West Africa. J. Clin. Microbiol. 38, 138–145 (2000).
Geisler, W.M., Suchland, R.J., Whittington, W.L. & Stamm, W.E. The relationship of serovar to clinical manifestations of urogenital Chlamydia trachomatis infection. Sex. Transm. Dis. 30, 160–165 (2003).
Gao, X. et al. Distribution study of Chlamydia trachomatis serovars among high-risk women in China performed using PCR-restriction fragment length polymorphism genotyping. J. Clin. Microbiol. 45, 1185–1189 (2007).
van de Laar, M.J. et al. Differences in clinical manifestations of genital chlamydial infections related to serovars. Genitourin. Med. 72, 261–265 (1996).
Parkhill, J. et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404, 502–506 (2000).
Otto, T.D., Sanders, M., Berriman, M. & Newbold, C. Iterative Correction of Reference Nucleotides (iCORN) using second generation sequencing technology. Bioinformatics 26, 1704–1707 (2010).
Zerbino, D.R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Thomson, N.R. et al. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18, 161–171 (2008).
Chain, P.S. et al. Genomics. Genome project standards in a new era of sequencing. Science 326, 236–237 (2009).
Darling, A.E., Mau, B. & Perna, N.T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5, e11147 (2010).
Kurtz, S. & Schleiermacher, C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics 15, 426–427 (1999).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Acknowledgements
We thank the core informatics, library-making and sequencing teams at the Wellcome Trust Sanger Institute. S.R.H. is grateful for the opportunity to discuss this project at the Permafrost conference. This work was funded by the Wellcome Trust grant numbers 098051 and 080348. B.G.S. was funded by the Wellcome Trust.
Author information
Authors and Affiliations
Contributions
S.R.H. assembled, aligned and analyzed the data and wrote the paper. I.N.C. jointly conceived of the project with N.R.T. and provided samples. H.M.B.S.-S. performed experiments, carried out analyses of the data and helped write the paper. L.T.C., P.M., R.J.S., M.J.H., D.M., R.W.P., D.A.L., M.U., K.P., C.B., R.B., H.J.C.d.V., S.A.M., A.W.S., C.M.B., A.S., M.C. and B.d.B. collected and cultured samples. B.G.S. and J.P. helped interpret the data and write the paper. N.R.T. conceived of and ran the project and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Tables 1 and 2. (PDF 2226 kb)
Rights and permissions
About this article
Cite this article
Harris, S., Clarke, I., Seth-Smith, H. et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet 44, 413–419 (2012). https://doi.org/10.1038/ng.2214
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2214
This article is cited by
-
A retrospective evaluation of the Euroarray STI-11 multiplex system for the detection of eight STI causing agents
Scientific Reports (2023)
-
Trachoma
Nature Reviews Disease Primers (2022)
-
High-resolution multilocus sequence typing for Chlamydia trachomatis: improved results for clinical samples with low amounts of C. trachomatis DNA
BMC Microbiology (2021)
-
Diagnosis of Chlamydia trachomatis genital infections in the era of genomic medicine
Brazilian Journal of Microbiology (2021)
-
Sensitivity, specificity, inclusivity and exclusivity of the updated Aptima Combo 2 assay, which provides detection coverage of the new diagnostic-escape Chlamydia trachomatis variants
BMC Infectious Diseases (2020)