Abstract
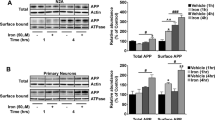
The recycling of the amyloid precursor protein (APP) from the cell surface via the endocytic pathways plays a key role in the generation of amyloid β peptide (Aβ) in Alzheimer disease. We report here that inherited variants in the SORL1 neuronal sorting receptor are associated with late-onset Alzheimer disease. These variants, which occur in at least two different clusters of intronic sequences within the SORL1 gene (also known as LR11 or SORLA) may regulate tissue-specific expression of SORL1. We also show that SORL1 directs trafficking of APP into recycling pathways and that when SORL1 is underexpressed, APP is sorted into Aβ-generating compartments. These data suggest that inherited or acquired changes in SORL1 expression or function are mechanistically involved in causing Alzheimer disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Mattson, M.P. Pathways towards and away from Alzheimer's disease. Nature 430, 631–639 (2004).
Goate, A. et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer disease. Nature 349, 704–706 (1991).
Sherrington, R. et al. Cloning of a gene bearing missense mutations in early onset familial Alzheimer's disease. Nature 375, 754–760 (1995).
Rogaev, E.I. et al. Familial Alzheimer's disease in kindreds with missense mutations in a novel gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature 376, 775–778 (1995).
Saunders, A.M. et al. Association of apoliprotein E allele e4 with the late-onset familial and sporadic Alzheimer disease. Neurology 43, 1467–1472 (1993).
Bales, K.R. et al. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat. Genet. 17, 263–264 (1997).
Golde, T.E., Estus, S., Younkin, L.H., Selkoe, D.J. & Younkin, S.G. Processing of the amyloid protein precursor to potentially amyloidogenic derivatives. Science 255, 728–730 (1992).
Haass, C. & Selkoe, D.J. Cellular processing of beta-amyloid precursor protein and the genesis of amyloid beta-peptide. Cell 75, 1039–1042 (1993).
Bayer, T.A. et al. Key factors in Alzheimer's disease: beta-amyloid precursor protein processing, metabolism and intraneuronal transport. Brain Pathol. 11, 1–11 (2001).
Kinoshita, A. et al. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J. Cell Sci. 116, 3339–3346 (2003).
Vetrivel, K.S. et al. Spatial segregation of gamma -secretase and substrates in distinct membrane domains. J. Biol. Chem. 280, 25892–25900 (2005).
Scherzer, C.R. et al. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch. Neurol. 61, 1200–1205 (2004).
Small, S.A. et al. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann. Neurol. 58, 909–919 (2005).
Andersen, O.M. et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA 102, 13461–13466 (2005).
Offe, K. et al. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J. Neurosci. 26, 1596–1603 (2006).
Pritchard, J.K. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 69, 124–137 (2001).
Pritchard, J.K. & Cox, N.J. The allelic architecture of human disease genes: common disease-common variant...or not? Hum. Mol. Genet. 11, 2417–2423 (2002).
Athan, E.S. et al. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. J. Am. Med. Assoc. 286, 2257–2263 (2001).
Bowirrat, A., Treves, T.A., Friedland, R.P. & Korczyn, A.D. Prevalence of Alzheimer's type dementia in an elderly Arab population. Eur. J. Neurol. 8, 119–123 (2001).
Rogaeva, E.A. et al. An alpha-2-macroglobulin insertion-deletion polymorphism in Alzheimer's disease. Nat. Genet. 22, 19–22 (1999).
Rogaeva, E. et al. Evidence for an Alzheimer disease susceptibility locus on chr 12, and for further locus heterogeneity. JAMA 280, 614–618 (1998).
Lee, J.H. et al. Fine mapping of 10q and 18q for familial Alzheimer's disease in Caribbean Hispanics. Mol. Psychiatry 9, 1042–1051 (2004).
Graff-Radford, N.R. et al. Association between apolipoprotein E genotype and Alzheimer disease in African American subjects. Arch. Neurol. 59, 594–600 (2002).
Green, R.C. et al. Risk of dementia among white and African American relatives of patients with Alzheimer disease. JAMA 287, 329–336 (2002).
Farrer, L.A. et al. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum. Mol. Genet. 12, 415–422 (2003).
Lin, S., Chakravarti, A. & Cutler, D.J. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat. Genet. 36, 1181–1188 (2004).
Ertekin-Taner, N. et al. Elevated amyloid beta protein (Abeta42) and late onset Alzheimer's disease are associated with single nucleotide polymorphisms in the urokinase-type plasminogen activator gene. Hum. Mol. Genet. 14, 447–460 (2005).
Ertekin-Taner, N. et al. Genetic variants in a haplotype block spanning IDE are significantly associated with plasma Abeta42 levels and risk for Alzheimer disease. Hum. Mutat. 23, 334–342 (2004).
Dodson, S.E. et al. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J. Neuropathol. Exp. Neurol. 65, 866–872 (2006).
Seaman, M.N. Recycle your receptors with retromer. Trends Cell Biol. 15, 68–75 (2005).
He, X., Li, F., Chang, W.P. & Tang, J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE). J. Biol. Chem. 280, 11696–11703 (2005).
Farrer, L.A. et al. Effects of age, sex, and ethnicity on the asscoiation between apolipoprotein E genotype and Alzheimer's Disease. A meta-analysis. APOE and Alzheimer's Disease Meta Analysis Consortium. JAMA 278, 1349–1356 (1997).
Rabinowitz, D. & Laird, N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum. Hered. 50, 211–223 (2000).
Vermeire, S. & Rutgeerts, P. Current status of genetics research in inflammatory bowel disease. Genes Immun. 6, 637–645 (2005).
Owen, M.J., Craddock, N. & O'Donovan, M.C. Schizophrenia: genes at last? Trends Genet. 21, 518–525 (2005).
Horvath, S. et al. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet. Epidemiol. 26, 61–69 (2004).
Lange, C., DeMeo, D., Silverman, E.K., Weiss, S.T. & Laird, N.M. PBAT: tools for family-based association studies. Am. J. Hum. Genet. 74, 367–369 (2004).
Lange, C., Silverman, E.K., Xu, X., Weiss, S.T. & Laird, N.M. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics 4, 195–206 (2003).
Lange, C. & Laird, N.M. Power calculations for a general class of family-based association tests: dichotomous traits. Am. J. Hum. Genet. 71, 575–584 (2002).
Van Steen, K. et al. Genomic screening and replication using the same data set in family-based association testing. Nat. Genet. 37, 683–691 (2005).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Jannot, A.S., Essioux, L., Reese, M.G. & Clerget-Darpoux, F. Improved use of SNP information to detect the role of genes. Genet. Epidemiol. 25, 158–167 (2003).
Terwilliger, J.D. & Weiss, K.M. Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr. Opin. Biotechnol. 9, 578–594 (1998).
Schaid, D.J., Rowland, C.M., Tines, D.E., Jacobson, R.M. & Poland, G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 70, 425–434 (2002).
Chen, F. et al. Nicastrin binds to membrane-tethered Notch. Nat. Cell Biol. 3, 751–754 (2001).
Benjannet, S. et al. Post-translational processing of beta-secretase (beta-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-beta production. J. Biol. Chem. 276, 10879–10887 (2001).
Yu, G. et al. A novel protein (nicastrin) modulates presenilin-mediated Notch/Glp1 and betaAPP processing. Nature 407, 48–54 (2000).
Gu, Y. et al. The presenilin proteins are components of multiple membrane-bound complexes which have different biological activities. J. Biol. Chem. 279, 31329–31336 (2004).
Hasegawa, H. et al. Both the sequence and length of the C terminus of PEN-2 are critical for intermolecular interactions and function of presenilin complexes. J. Biol. Chem. 279, 46455–46463 (2004).
Acknowledgements
The authors thank J. Ott for advice on the statistical analysis. The authors acknowledge the work of B. Tycko, M. Medarano, R. Lantigua, Y. Stern, A. Akomolafe, J. Browndyke, H. Chui, R. Go, A. Kurz, H. Petrovitch, N. Relkin, D. Sadovnick, P. Erlich, S. Sunyaev, L. Ma, J. Lok and S. Younkin. This work was supported by the Canadian Institutes of Health Research, the Howard Hughes Medical Institute, the Canadian Institutes of Health Research–Japan Science and Technology Trust, the Alzheimer Society of Ontario, the Canada Foundation for Innovation, the Ontario Research and Development Challenge Fund, the Ontario Mental Health Foundation, Genome Canada, the US National Institutes of Health and the National Institute on Aging (grants R37-AG15473 and P01-AG07232 (R.M.), R01-AG09029 (L.A.F.), RO1-HG/AG02213 (R.C.G.), P30-AG13846 (L.A.F., R.C.G.), R01-AG017173 (R.P.F., L.A.F.), P50-AG16574 (R.C.P., S.Y., N.G.R.) and U01-AG06786 (R.C.P.)), the Alzheimer Association, the Alzheimer Society of Canada, the Blanchett Hooker Rockefeller Foundation, the Charles S. Robertson Gift (R.M.), Fonds de la Recherche en Santé (Y.M.), Assessorato Regionale alla Sanità-Regione Calabria, Finalized Project of the Ministry of Health (2003–2005) (A.C.B.), Fondation pour la Recherche Médical, Robert and Clarice Smith and Abigail Van Buren, the Alzheimer Disease Research Program (R.P., S.Y.) and the W. Garfield Weston Fellowship (E.R., G.S.U.).
Author information
Authors and Affiliations
Contributions
The authors' roles are described in Supplementary Table 11 online.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Characteristics of genotyped subjects. (PDF 17 kb)
Supplementary Table 2
Single SNP results for VPS10 genes other than SORL1. (PDF 27 kb)
Supplementary Table 3
Characteristics of SNPs in SORL1. (PDF 579 kb)
Supplementary Table 4
Single SNP results for all 29 SORL1 SNPs in the six primary datasets. (PDF 355 kb)
Supplementary Table 5
Three-SNP haplotypes for all SORL1 SNPs. (PDF 245 kb)
Supplementary Table 6
Two-SNP haplotypes for all SORL1 SNPs. (PDF 232 kb)
Supplementary Table 7
Four-SNP haplotypes for all SORL1 SNPs. (PDF 284 kb)
Supplementary Table 8
Five-SNP haplotypes for all SORL1 SNPs. (PDF 261 kb)
Supplementary Table 9
Six-SNP haplotypes for all SORL1 SNPs. (PDF 372 kb)
Supplementary Table 10
Rare sequence variants in SORL1. (PDF 65 kb)
Supplementary Table 11
Authors' roles in the study. (PDF 21 kb)
Rights and permissions
About this article
Cite this article
Rogaeva, E., Meng, Y., Lee, J. et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet 39, 168–177 (2007). https://doi.org/10.1038/ng1943
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1943
This article is cited by
-
A familial missense variant in the Alzheimer’s disease gene SORL1 impairs its maturation and endosomal sorting
Acta Neuropathologica (2024)
-
LRP10 and α-synuclein transmission in Lewy body diseases
Cellular and Molecular Life Sciences (2024)
-
Step by step: towards a better understanding of the genetic architecture of Alzheimer’s disease
Molecular Psychiatry (2023)
-
On the causal role of retromer-dependent endosomal recycling in Alzheimer’s disease
Nature Cell Biology (2023)
-
The Role of RIN3 Gene in Alzheimer’s Disease Pathogenesis: a Comprehensive Review
Molecular Neurobiology (2023)